CeMM - Research Center for Molecular Medicine, Austrian Academy of Sciences,
Ludwig Boltzmann Institute for Network Medicine, University of Vienna
Vienna, Austria

Research highlights
Spatially resolved biology and tissue architecture:
- Buljan, et al, BioRxiv, 2025
- Abila, Buljan, Zheng, et al., BioRxiv, 2024
- Kim, et al., Nature Methods, 2022
Computational methods and software:
Latest news
2025-12-06 - StainX: CUDA-optimized stain normalization is now open source!
2025-11-21 - Ernesto presented his work at Cytodata 2025
2025-11-20 - Andre gave a talk at the 2nd European CyTOF conference
2025-11-19 - LazySlide was showcased at the 2025 scverse conference
2025-11-03 - Samir joins the lab!
2025-10-31 - We joined the 2025 CeMM halloween party!
More news here.We are a research group at CeMM - the Research Center for Molecular Medicine of the Austrian Academy of Sciences and at the Ludwig Boltzmann Institute for Network Medicine at the University of Vienna working on computational and molecular methods to study human aging and pathology.

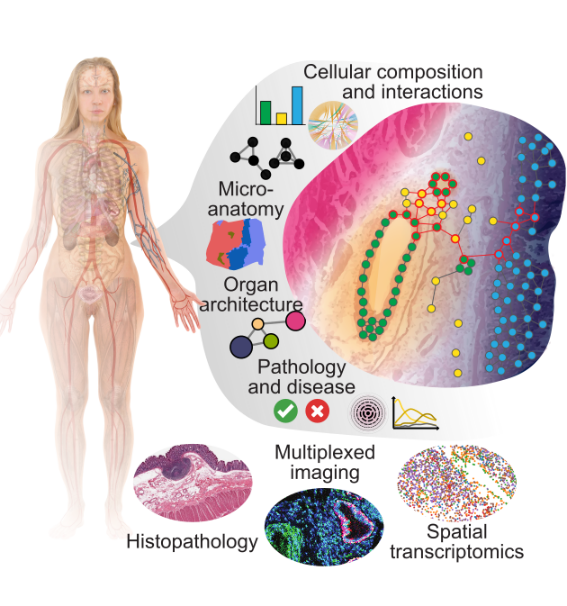
We are interested in uncovering the architectural patterns of the human body across anatomical scales: cellular and micro-anatomical organization, and organ-wide patterns.
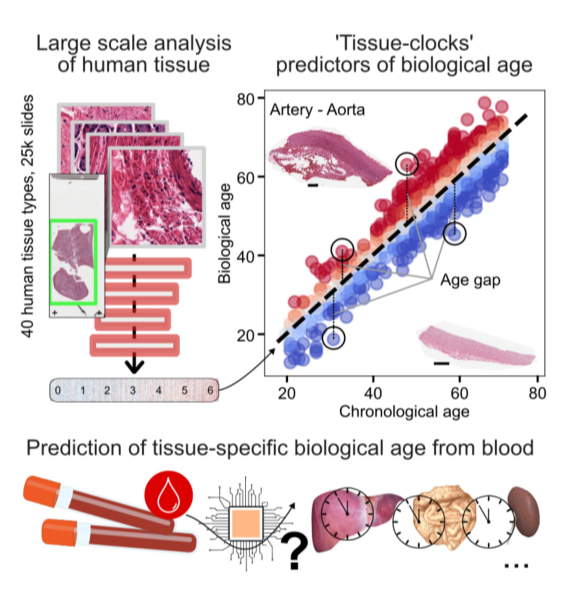
We think that these patterns hold the key to understand how cellular changes lead to the progressive loss of physiological ability as individuals age and can be predictive of disease risk and its onset.
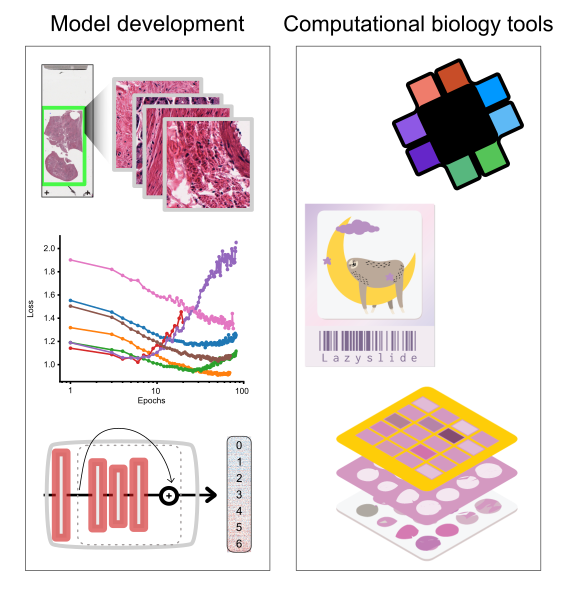
To do that, we primarily develop computational methods and software tools for the analysis of spatial data (digital pathology, spatial transcriptomics, highly multiplexed imaging), and its integration with various modalities of molecular and clinical data of individuals along their lifespan.